Identification of Predictive ERBB Mutations by Leveraging Publicly Available Cell Line Databases
Abstract
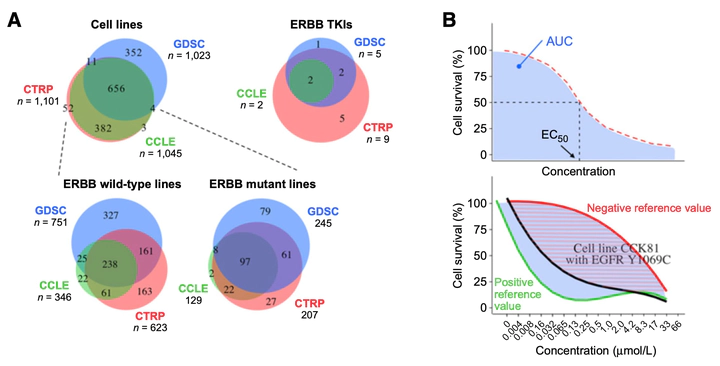
While targeted therapies can be effective for a subgroup of patients, identification of individuals who benefit from the treatments is challenging. At the same time, the predictive significance of the vast majority of the thousands of mutations observed in the cancer tissues remains unknown. Here, we describe the identification of novel predictive biomarkers for ERBB-targeted tyrosine kinase inhibitors (TKI) by leveraging the genetic and drug screening data available in the public cell line databases: Cancer Cell Line Encyclopedia (CCLE), Genomics of Drug Sensitivity in Cancer (GDSC), and Cancer Therapeutics Response Portal (CTRP). We assessed the potential of 412 ERBB mutations in 296 cell lines to predict responses to 10 different ERBB-targeted TKIs. Seventy-six ERBB mutations were identified that were associated with ERBB TKI sensitivity comparable to non-small cell lung cancer cell lines harboring the well-established predictive EGFR L858R mutation or exon 19 deletions. Fourteen (18.4 %) of these mutations were classified as oncogenic by the cBioPortal database, whereas 62 (81.6 %) were regarded as novel potentially predictive mutations. Out of nine functionally validated novel mutations, EGFR Y1069C and ERBB2 E936K were transforming in Ba/F3 cells and demonstrated enhanced signaling activity. Mechanistically, the EGFR Y1069C mutation disrupted the binding of the ubiquitin ligase c-CBL to EGFR, whereas the ERBB2 E936K mutation selectively enhanced the activity of ERBB heterodimers. These findings indicate that integrating data from publicly available cell line databases can be used to identify novel, predictive non-hotspot mutations, potentially expanding the patient population benefiting from existing cancer therapies.